MetaSanity - Output
Published:
In a previous post, we walked through several examples of using BioMetaDB - a Bio-focused command-line SQL wrapper package - to query the results of our MetaSanity pipeline runs. The contents of each project is self-contained, making it very easy for users to focus their attention on getting specific information.
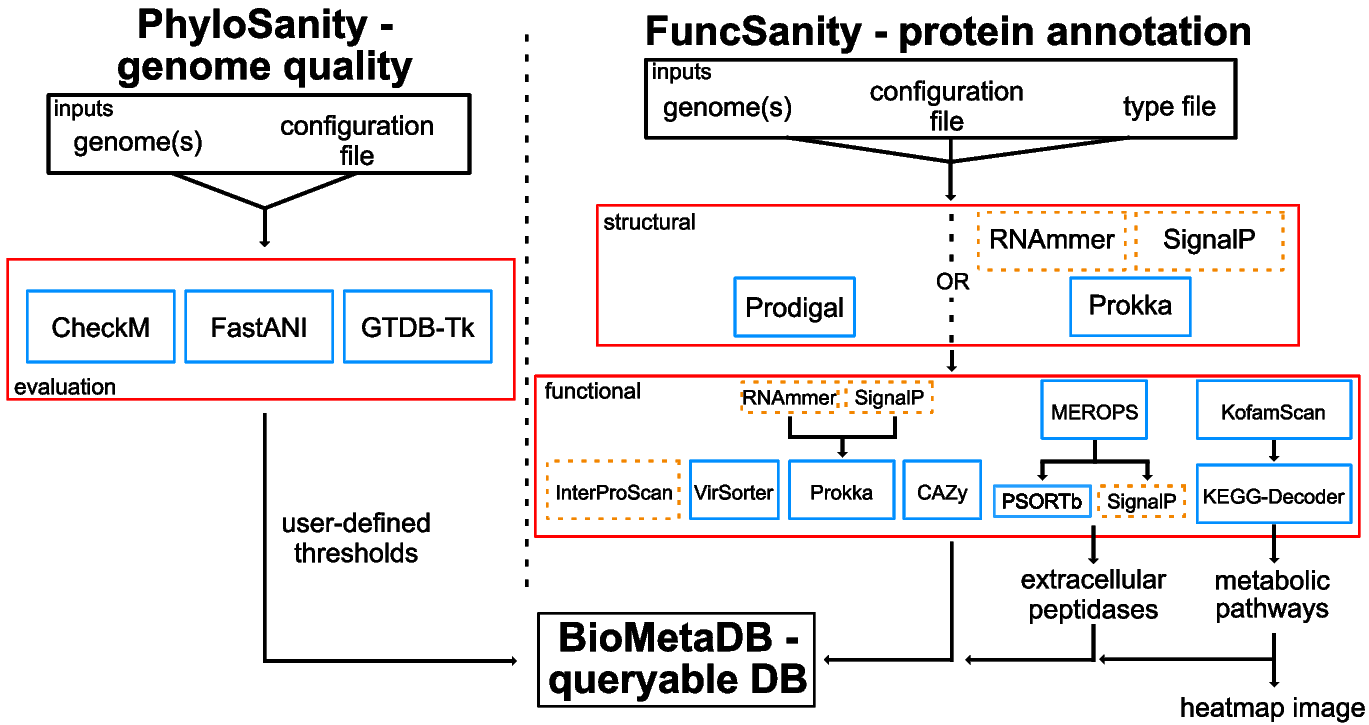
Given the complexity and volume of this application, we will walk through the raw and parsed output from each of the programs that MetaSanity runs. This information is also available as a PowerPoint presentation (views best using PowerPoint Online).
MetaSanity output
out/
├── evaluation.tsv
├── functions.tsv
├── checkm_results/
├── fastani_results/
├── gtdbtk_results/
├── prodigal_results/
├── prokka_results/
├── interproscan_results/
├── kegg_results/
├── cazymerops_results/
├── virsorter_results/
├── mag1.annotation.tsv
...The first two .tsv files contain the parsed results of the MetaSanity pipelines. These are simple tab-delimited text files. PhyloSanity results are contained in evaluation.tsv. FuncSanity results are in a set of files. functions.tsv contains functional and metabolic information for all genomes that were studies. A file ending with the extension .annotation.tsv will be generated for each genome that was analyzed.
The remaining directories contain the raw output from each of the programs that were incorporated into the user’s annotation pipeline. The resulting file(s) are parsed for specific information to generate the workflow’s results.
PhyloSanity
CheckM
checkm_results/checkm_lineageWF_results.qa.txt is parsed for contamination and completion estimates.
FastANI
fastani_results/fastani_results.txt contains ANI information used in redundancy determination
GTDB-Tk
gtdbtk_results/GTDBTK.bac120.summary.tsv and gtdbtk_results/GTDBTK.ar122.summary.tsv optionally provide putative phylogeny.
FuncSanity
For each MAG that is analyzed, a series or files may be generated.
Prodigal gene caller
prodigal_results/
├── mag1.mrna.fna
├── mag1.protein.faa
├── mag1.txtProkka (optional gene caller and annotation pipeline)
prokka_results/
├── mag1/
├── mag1.tsv # Original output
├── mag1.prk.tsv.amd # Parsed output for BioMetaDB
├── mag1.prk.tsv.prokka.nucl # RNA features
├── diamond/
├── mag1.prk-to-prd.tsv # Protein annotationsThe remaining files in each directory are the outputs from each Prokka run.
InterProScan
interproscan_results/
├── mag1.tsv # Original output
├── mag1.amended.tsv # Parsed outputInterProScan contributes the most to MetaSanity runtimes.
KoFamScan and KEGG-Decoder
kegg_results/
├── biodata_results/ # KEGG-Decoder-derived metabolic pathway estimation counts
├── KEGG.final.tsv
├── KEGG.decoder.tsv
├── combined_results/
├── combined.ko # kofamscan matches for KEGG-Decoder input
├── combined.protein # gene calls for KEGG-Decoder input
├── combined.expander.tbl # HMM search results (part of BioData pipeline)
├── combined.expander.tsv # Raw counts from BioData pipeline
├── combined.html # Heatmap of putative metabolic pathway completion estimates
├── kofamscan_results/
├── mag1.tsv # KO matches per protein
├── mag1.detailed # Detailed kofamscan output
├── mag1.amended.tbl # Parsed output for BioMetaDBMEROPS
cazymerops_results/
├── mag1.merops.tsv # MEROPS matches
├── mag1.pfam.tsv # MEROPS matches by PFam id
├── merops/
├── mag1.merops.protein.faa # MEROPS protein matches
├── hmmconvert_data/ # HMM prep
├── hmmsearch_results/ # HMMSearch outputCAZy
cazymerops_results/
├── cazy/
├── hmmsearch_results/ # HMMSearch output
├── mag1.cazy_assignments.tsv # Raw counts of CAZy HMM matches
├── mag1.cazy_assignments.byprot.tsv # Protein annotationsPSORTb and SignalP
cazymerops_results/
├── psortb_results/
├── mag1.tbl # Raw PSORTb output
├── signalp_results/
├── mag1.signalp.tbl # Raw SignalP outputExtracellular Peptidase
cazymerops_results/
├── mag1.pfam.by_prot.tsv # PFam annotations for peptidase resultsVirSorter
virsorter_results/
├── mag1
├── virsorter_results/
├── VIRSorter_global-phage-signal.csv # Original output
├── mag1.VIRSorter_adj_out.tsv # Parsed output for BioMetaDBThe remaining files in each virsorter_results directory are the raw outputs of the VirSorter run for each MAG.
